11. Why is a new population of people exhibiting disease symptoms?
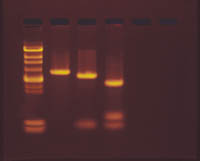
Gel courtesy of Edvotek (www.edvotek.com)
In this lab you get to incorporate what you have learned in lecture (Family Reunion) and labs (9., 10.). In Lab 9. you investigated how bacteria can acquire immunity to antibiotics naturally through mutation of their DNA. In Lab 10, you had the opportunity to investigate the use of recombinant DNA technology to transfer genetic material from one organism into another using plasmids. This resulted in the bacteria acquiring new characteristics. Now you are investigating the DNA itself.
In Family Reunion, you learned that each chromosome is a molecule of DNA. DNA is composed of a sequence of nucleotides linked together into a twisted ladder structure. The rungs of the ladder are formed by links between "base-pairs" of the chemcials Adenine-Thymine or Cytosine-Guanine. Along each side of the molecule, these bases form a sequence that represents the genetic code for a protein. There is actually a lot more there than just the code for proteins, but this is unneccessary for us for now. Each set of three bases (a triplet or codon) identifies which amino acid should be connected next in the protein. Certain codons signify where the information starts or stops. The sequence of DNA that codes for a particular protein (polypeptide or amino acid sequence) can be referred to as a gene. A change in the DNA results in a change in the amino acid sequence and can result in a change in the structure of the protein and thus its ability to perform a particular function. The process of protein synthesis also involves copying the DNA code as a mRNA sequence during transcription and the bonding of the amino acids brought to the ribosomes by the tRNA molecules that bear the matching RNA sequence (anti-codon) to the one on the mRNA during the process of translation.
Mutations are changes in the DNA. They can be a result of point mutations (substitutions, deletions, or insertions of single bases) or restructuring of chromosomes (loss of a fragment, addition of a fragment, inversion of a portion). Such changes result in one molecule of DNA (chromosome) being different from another, even from its homologue. Sequencing a strand of DNA (determining the order of the nucleotides(A,T,C,G) can be done to compare DNA molecules, but this is time consuming and tedious. Other methods can let a scientist determining if two pieces of DNA are the same more quickly. Digesting DNA with restriction enzymes and analyzing the DNA using electrophoresis is one technique.
When you transformed bacteria in Lab 10, you inserted plasmids into them. These circular pieces of DNA contain a few genes on them. The bacteria that picked up the plasmid took on certain characteristics, reproduced, and grew into noticeable colonies. What allowed us to create the plasmids with the genes in which we were interested was the use of restriction enzymes which cut up the DNA at certain places. DNA containing the gene of interest was cut with the same restriction enzyme. The resulting pieces are then allowed to join form recombinant DNA. You should be able to explain how that results in the fluorescent colonies you saw. You should also be able to explain how the non-fluorescent DNA described in lab 10 failed to produce the same type of bacteria and how antibiotic resistance was controlled or acquired in these bacteria.
Now if two pieces of DNA, in this case the plasmids, contain different mutations (DNA sequences), how can we detect the differences in the DNA itself? To understand how, you need to review/understand what restriction enzymes do. The prelab from Investigation 10 included three tutorials and a reading that explained DNA structure and how restriction enzymes cut the DNA at specific sequences. They were used to create the DNA (plasmids) you used in Investigation 10. They can also be used to cut the DNA into pieces, with each piece being a different length. Mutations that result in different DNA sequences, can result in fragments of different lengths (Can you explain how?) when cut with the same restriction enzyme. We can detect these different length fragments using gel electrophoresis. The pre-lab exercises for Investigation 10 explained this technique. Some of the websites below explain it also. The prelab for this investigation will show you how to and give you a chance to practice preparing a gel.